Data / Code
Software
Much Schwilk lab software and data is available through our GitHub organization page. Dylan Schwilk’s projects can also be found on his GitHub page. Some repositories of possible interest: a student Emacs configuration, a collection of pandoc templates organized into a build system, and our protocols repo.
Online data and source code for published papers
Below, in reverse chronological order, I have listed publications for which I have posted source code and/or data.
Schwilk et al, 2013 PLoS One
BarkContour.R: R script to read digitized bark contours and calculate bark thickness distributions.
Morlon et al, 2011, Ecology Letters
dwstree project at github: Source python code for phylogenetic analysis including generalized version of the BLADJ node age assignment algorithm. The main script of interest should be rarefaction.py which provides the sensitivity analyses and makes use of the branch length assignment in branch_lengths.py. Rarefaction code is written to perform trivial parallelization on multi-node computers. The phylomatic algorithm with ability to maintain the full phylogeny with non species-level tips is found in treematic.py. Further documentation is in source code docstrings.
Cornwell, W.K, D.W. Schwilk and D.D. Ackerly. 2006. A trait-based test for habitat filtering: convex hull volume. Ecology 87: 1465–1471.
traithull Provides an interface to the Qhull program that allows ecologists to easily calculate the convex hull volume (CHV) metric of functional diversity and do tests against null models. For a description of the method see: Cornwell, W.K, D.W. Schwilk and D.D. Ackerly. 2006. A trait-based test for habitat filtering: convex hull volume. Ecology 87: 1465–1471.
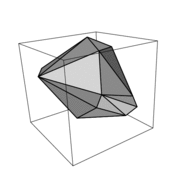
Schwilk, D. W. and D. D. Ackerly. 2005. Limiting similarity and functional diversity along environmental gradients. Ecology Letters 8:272-281.
The source C++ code for the niche and limiting similarity simulation (kniche), and the for the numerical model (aniche) as well as python scripts used to generate parameter inputs are all available: ms-data-EcologyLetters-2005
Schwilk, D. W. and B. Kerr. 2002. An alternative explanation for the evolution of flammability. Oikos 99: 431-442.
The C++ code for the genetic “niche-hiking” simulation: ms-data-Oikos-2002
Schwilk, D. W., and D. D. Ackerly. 2001. Flammability and serotiny as strategies: correlated evolution in pines. Oikos 94: 326-336.
Online appendix with supertree phylogenies created and used in above paper: ms-data-Oikos-2001